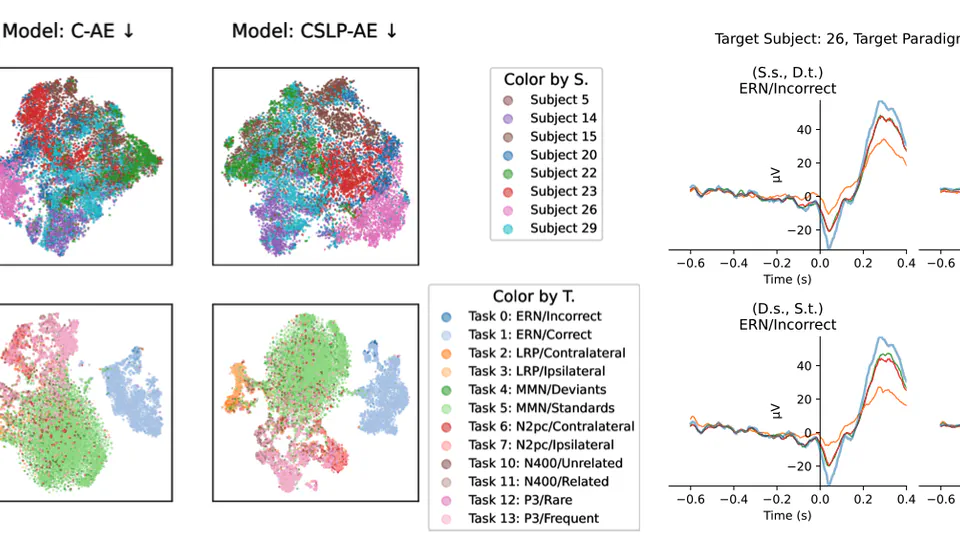
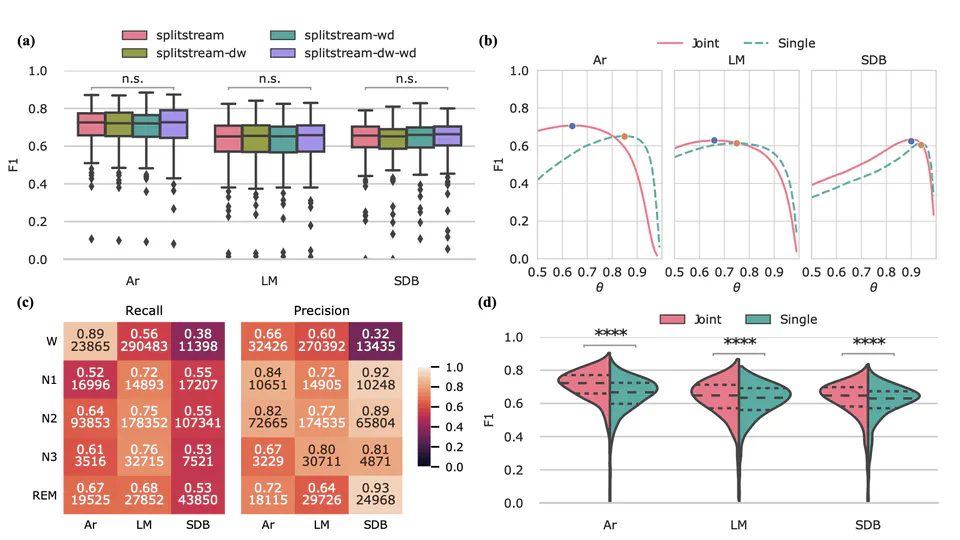
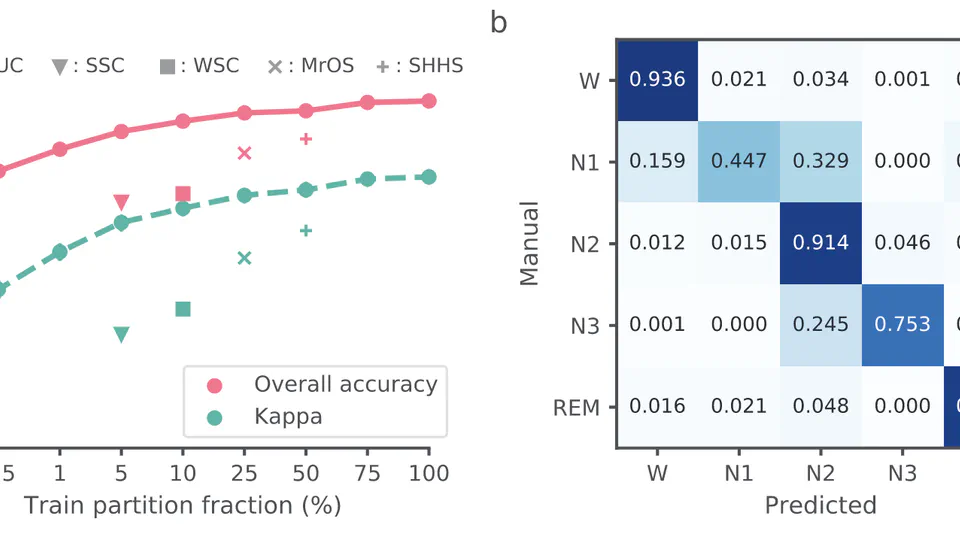
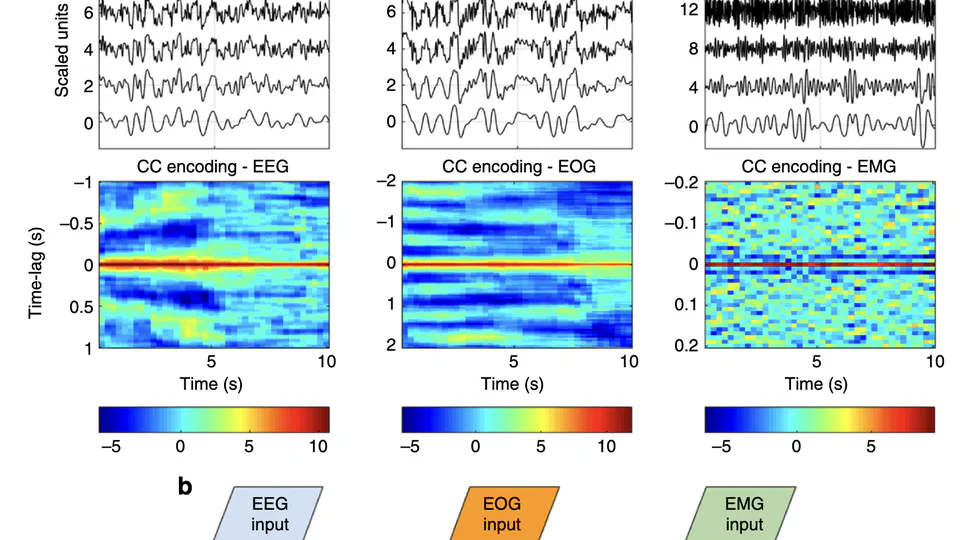
I am a research scientist and biomedical engineer with special interests in signal processing and machine learning for computational sleep science. I design algorithms and data processing pipelines for modeling, analysis and visualisation of physiological signals such as EEG, EOG and EMG obtained from nocturnal PSG recordings. Through this work, I hope to further our collective understanding of sleep pathologies, and how they impact human health.
Currently, I work as a postdoctoral researcher at the Section for Cognitive Systems at the Department of Applied Mathematics and Computer Science, Technical University of Denmark, through a LF Postdoc grant from the Lundbeck Foundation.
I also provide consulting services to academic and industry research groups, which include data processing and machine learning pipelines, algorithm development, and data visualization.
In 2020, I graduated with a PhD in Biomedical Engineering from the Technical University of Denmark under the supervision of the late Associate Professor Helge B. D. Sørensen, PhD; Professor Poul Jennum, MD, PhD, from the Danish Center for Sleep Medicine; and Professor Emmanuel Mignot, MD, PhD, from Stanford University.
- Deep learning
- Computational sleep science
- Biomedical signal processing
PhD in Biomedical Engineering
Technical University of Denmark
MScEng in Biomedical Engineering
Technical University of Denmark
BScEng in Biomedical Engineering
Technical University of Denmark